The Jaccard similarity compares two sets of data to determine how similar they are. The value of the Jaccard similarity can be between 0 and 1, where the close the number is to 1 the more similar the two sets of data are.
This tutorial will go through how to calculate the Jaccard similarity using R with code examples.
Table of contents
Jaccard Similarity Formula

The Jaccard Similarity is a term coined by Paul Jaccard, defined as the size of the intersection divided by the size of the union of two sets. In simple terms, we can determine the Jaccard Similarity as the number of objects the two sets have in common divided by the total number of objects. If two datasets share the same members, the Similarity term will be 1. Conversely, if the two sets have no members in common, then the term will be 0.
Jaccard Similarity in R
We can define a similarity function which takes two vectors as input.
jacc <- function(x, y) {
intersection = length(intersect(x, y))
union = length(x) + length(y) - intersection
return (intersection/union)
}
The Jaccard Similarity (J) is (the count of common elements in both sets) / ( the count of elements of the first set plus the count of elements in the second set minus the common elements in both sets)
Jaccard Similarity in R For Numeric Sets
Consider an example with two numeric vectors. We will call the user-defined function as follows:
x <- c(2, 3, 5, 6, 7, 11, 19, 21, 7, 4) y <- c(19, 11, 4, 8, 22, 1, 3, 10, 17, 21) jacc(x,y)
[1] 0.3333333
Jaccard Similarity in R For Character Vectors
We can also use the function for sets containing strings. Consider an example of two character vectors. We will call the user-defined function as follows:
x <- c('london', 'paris', 'tokyo', 'berlin', 'accra')
y <- c('accra', 'cairo', 'geneva', 'tokyo', 'lusaka')
jacc(x, y)
[1] 0.25
The Jaccard Distance
The Jaccard distance measures the dissimilarity between sets, is complementary to the Jaccard Similarity, and is obtained by subtracting the Jaccard coefficient from 1, or equivalently by dividing the difference of the size of the union and the intersection of two sets by the size of the union:
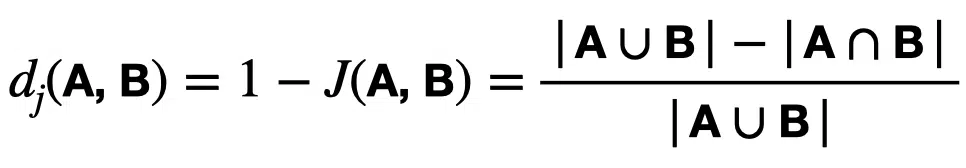
The distance is a metric on the collection of all finite sets. We can use the distance to calculate an n $latex \times$ n matrix for clustering and multidimensional scaling of n sample sets.
Jaccard Distance In R for Numeric Vectors
We can use the previously defined function to determine the Jaccard distance between two numeric sets, which is 1 – Jaccard similarity. Consider the following example with two numeric vectors:
x <- c(2, 3, 5, 6, 7, 11, 19, 21, 7, 4) y <- c(19, 11, 4, 8, 22, 1, 3, 10, 17, 21) jaccard_distance <- 1 - jacc(x,y) jaccard_distance
[1] 0.6666667
Jaccard Distance In R for Character Vectors
We can use the previously defined function to determine the Jaccard distance between two character sets, which is 1 – Jaccard similarity. Consider the following example with two character vectors:
x <- c('london', 'paris', 'tokyo', 'berlin', 'accra')
y <- c('accra', 'cairo', 'geneva', 'tokyo', 'lusaka')
jaccard_distance <- 1 - jacc(x, y)
jaccard_distance
[1] 0.75
Jaccard Similarity Matrix in R
We can calculate a Jaccard similarity matrix using the vegdist() function from the vegan package. Consider the following example of a data frame consisting of two numeric vectors, we will pass the data frame to the vegdist() method and set the method to “jaccard“.
install.packages("vegan")
library(vegan)
x <- c(2, 3, 5, 6, 7, 11, 19, 21, 7, 4)
y <- c(19, 11, 4, 8, 22, 1, 3, 10, 17, 21)
df <- data.frame(x, y)
vegdist(df, method="jaccard")
Let’s run the code to see the result:
1 2 3 4 5 6 7 8 9 2 0.4090909 3 0.7500000 0.5625000 4 0.6000000 0.3529412 0.3571429 5 0.2758621 0.5172414 0.6896552 0.5172414 6 0.9000000 0.8181818 0.6000000 0.6315789 0.7575758 7 0.8684211 0.8000000 0.6521739 0.6666667 0.7560976 0.4545455 8 0.7000000 0.5937500 0.7096774 0.5483871 0.6046512 0.6129032 0.2903226 9 0.2692308 0.4166667 0.6250000 0.4166667 0.1724138 0.7142857 0.7222222 0.5526316 10 0.1600000 0.4400000 0.6923077 0.5555556 0.1379310 0.8437500 0.8250000 0.6666667 0.2500000
Summary
Congratulations on reading to the end of this tutorial. Jaccard Similarity provides a simple and intuitive method of finding the similarity between objects.
For further reading on how to calculate the Jaccard similarity in Python, please visit our guide: Jaccard Similarity
If you want to learn how to calculate the Jaccard similarity in C++, please visit our guide: Set Similarity with Jaccard Coefficient and MinHash in C++.
If you want to learn how to implement string similarity metrics in Python, R, and C++, please visit our guide: Jaro-Winkler Similarity: A Comprehensive Guide to String Matching.
Have fun and happy researching!
Suf is a senior advisor in data science with deep expertise in Natural Language Processing, Complex Networks, and Anomaly Detection. Formerly a postdoctoral research fellow, he applied advanced physics techniques to tackle real-world, data-heavy industry challenges. Before that, he was a particle physicist at the ATLAS Experiment of the Large Hadron Collider. Now, he’s focused on bringing more fun and curiosity to the world of science and research online.

